Many protein structures are elucidated to atomic detail using x-ray crystallography or nuclear magnetic resonance (NMR) experiments and are deposited in the PROTEIN DATA BANK (PDB). However, macromolecular assemblies are most relevant in understanding biological processes. Cryo-electron microscopy (cryo-EM) is an experimental technique that generates 3D electron density maps of medium resolution (7-10Å) for large macromolecular assemblies. A fitting or docking procedure for known atomic models within the assembly is needed to place these models into the experimental density map. Initial likely placements are refined to find the local optimum and discard incorrect arrangements. In recent years, graphical processing units (GPUs) have grown in popularity since they can now be programmed with higher-level languages like the industry standard OpenCL. GPUs are ideal in processing parallel computations and show significant speed ups over CPU implementations. The presented work applies GPUs to rapidly refine initial placements of protein models into low resolution density maps thereby accelerating construction of atomic-detail models for large macromolecular assemblies.
The schematic protocol ist:


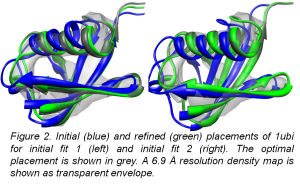

Alumni Project Members: Nils Woetzel, Edward W. Lowe Jr
